[AWC 1st Place] Epigenetic Reprogramming — The Elixir of Life?
- Admin

- Sep 18, 2023
- 30 min read
Look at this graph of life expectancy against time from 1770 up to 2021; notice how there’s a spike in life expectancy from 30 years in 1870 to 72 years in 2021. Within less than 2 centuries, the life expectancy of a human has more than doubled.

Figure 1: Graph showing human life expectancy from 1770 to 2021. Image taken from
Our World in Data.
This is all thanks to rapid advancements in medical research, with the discovery of antibiotics being arguably the largest breakthrough in longevity to date, extending human life expectancy by 23 years on average (Hutchings, Truman and Wilkinson, 2019). That being said, the increase in life expectancy has gradually slowed down, causing many to think that we may be approaching our biological limit. Regardless, researchers in the field of longevity have not given up and continue to churn out research suggesting we may be able to live longer than we thought.
Enter epigenetic reprogramming, a growing field that aims to “reverse” aging by ameliorating epigenetic changes in our cells.
What is aging?
Aging is a multifaceted biological process that manifests as a gradual decline in physiological function and homeostatic mechanisms with time. The process of aging holds significance as it is generally associated with risks of many diseases including cancer (White et al., 2014), metabolic disorders (Xia et al., 2021), cardiovascular diseases (North and Sinclair, 2012), and neurodegenerative diseases (Hou et al., 2019).
What is epigenetics?
Ever wondered why there are different cells in our body, although every cell in the body has the same set of genomes? Well, it’s because of epigenetics.
Our genome consists of tens of thousands of genes stored in the form of double-stranded deoxyribonucleic acid (DNA) in the nucleus of each cell. In order for a gene to be expressed, the DNA has to be transcribed into ribonucleic acid (RNA) and translated into proteins. DNA is usually coiled around a protein called histones, like a thread wound around a spool, to form chromatin (Quina, Buschbeck and Di Croce, 2006). The chromatin can exist as loose euchromatin, which can be accessed and facilitates gene expression, or dense heterochromatin, which is inaccessible and blocks gene expression (Li, Carey and Workman, 2007). Chemical alterations to histone proteins as well as DNA molecules can induce the formation of either euchromatin or heterochromatin. Chemical alterations to histone proteins include acetylation, methylation, phosphorylation, and ubiquitylation. Acetylation of histones is generally associated with the expression of genes (Bannister and Kouzarides, 2011), while methylation, phosphorylation, and ubiquitylation are less straightforward in their effects and could cause both repression and expression of genes (Al, Simpson and Ishwarlal Jialal, 2019). Chemical alterations of DNA molecules usually involve methylation of a cytosine nucleotide in a cytosine-guanine sequence. Methylated cytosines recruit gene suppressor proteins and reduce interaction between DNA and transcription factors (Moore, Le and Fan, 2012).
The difference in which genes are expressed or not determines the fate of a cell, whether it becomes a neuron, a muscle cell, or other lineages of cell types. The regulation of chemical alterations by various factors such as molecular signaling, lifestyle, environment, and disease states is known as epigenetics (Al, Simpson and Ishwarlal Jialal, 2019). Simply, epigenetics refers to the reversible process of altering which specific genes are expressed without any
underlying changes to the genome.

Figure 2: Graphical representation of the formation of chromatin from histones and DNA.
Image taken from the National Human Genome Research Institute.
Epigenetics and Aging
Although the exact causes of aging have not been pinpointed, continuous research has characterized the cellular and molecular hallmarks of aging. One of those hallmarks is epigenetic alterations. In young individuals, cells within each cell type have the same gene expression, due to having similar epigenetic information. However, as an individual ages, epigenetic changes occur spontaneously due to environmental and internal factors. This results in the formation of abnormal chromatin, characterized by different histone variants being incorporated, altered DNA methylation patterns, and altered histone modification patterns. Altered epigenetic information leads to different transcriptional patterns, and can also lead to genetic mutations (Pal and Tyler, 2016). As a result of the different transcriptional patterns and mutations, genes are expressed incorrectly. Cellular dysfunction may ensue due to the expression of certain genes, or the production of misfolded proteins (Douglas and Dillin, 2010).

Figure 3: Overview of epigenetic changes during aging. Image taken from Science
Advances.
Rewinding the clock of life
Despite the apparently unidirectional process of aging, the ability of the aging clock to be put on hold and reversed is fundamental to the nature of life. With every fertilization event, a zygote is formed, in the case of humans from the fusion between a sperm cell and an egg cell, both of which have chronological age measuring into the decades. Yet, the product of this noble merging is a cell that somehow erases any trace of aging. This reprogramming process is even more accentuated in experiments using somatic cell nucleus transfer, in which the nucleus of a somatic cell is transferred to an enucleated oocyte, such as the pioneering work of Dr. John Gurdon that showed for the first time that differentiated nuclei from tadpole intestinal or muscle cells could be transferred into enucleated Xenopus eggs and give rise to mature and fertile male and female frogs (Gurdon, 1962).

Figure 4: Prof. John B. Gurdon SCNT experiment. Image taken from Gurdon, 2013,
Nobel Lecture.
The fact that the nuclei were capable of giving rise to viable embryos that were themselves capable of developing into fertile adults and did not exhibit premature aging is evidence that the chronological age of the donor nuclei had been reset. The fact that somatic cells can be rejuvenated and have their pluripotency (ability to differentiate into any cell lineage) restored has opened the doors to many seeking to induce pluripotent states in cells by synthetic means.
Epigenetic reprogramming
Over the years, numerous research has sought to induce pluripotency, but one has risen as a major player in the field — epigenetic reprogramming. As cells differentiate and age, they accumulate epigenetic alterations that allow them to maintain their identity and remain stable. The theory is that by reversing the epigenetic alterations, we are able to revert cells into their primordial states, forming induced pluripotent stem cells (iPSCs).
Pioneering work was done back in 2006 by Yamanaka and Takahashi whereby they discovered the overexpression of four transcription factors: Oct4, Sox2, Klf4, and c-Myc, now known as “Yamanaka factors” or “OSKM, rearranges the epigenetic landscape, and converts somatic cells to a pluripotent state (Takahashi and Yamanaka, 2006). This research had such a large impact that Shinya Yamanaka received the Nobel Prize in Physiology or Medicine in 2012.
In the original study, it was shown that Oct4 and Sox2 were responsible for initiating the pluripotent state, while Klf4 and c-Myc were essential for reprogramming efficiency (Takahashi and Yamanaka, 2006). Global analysis of gene expression during reprogramming suggests that the process occurs in a stepwise manner (Mah et al., 2011), where the initial stage involves enhanced expression of genes that control DNA replication and cell division, along with repressed expression of genes responsible for cell adhesion and cell-cell contact. Single-cell expression analysis revealed that in fibroblasts, exogenous OSKM randomly trigger a cascade of events leading to the formation of a small proportion of pre-IPSCs. The expression of genes during that cascade of events in turn activates predictive markers of reprogramming — oestrogen-related receptor beta (Esrrb1) and Utf1. This is a key step that activates endogenous Sox2 (Polo et al., 2012). Once endogenous Sox2 is activated, Sox2-dependent activation of SALL4 and LIN28 activates the expression of genes associated with pluripotency such as fibroblast growth factor 4 (FGF4), F-box protein 15 (FBXO15), and DNA cytosine-5-methyltransferase 3β (DNMT3B). This, in turn, leads to the activation of endogenous Oct3/4 and NANOG, which is a critical factor for the maintenance of the undifferentiated state of pluripotent cells (Buganim et al., 2012).
One of the Yamanaka factors, c-Myc activates pluripotency markers and combines with histone acetyltransferase complexes to induce global histone acetylation, opening up the chromatin for binding of transcription factors, which in this case is exogenous Oct4 and Sox2 (Soufi, Donahue and Zaret, 2012). Another Yamanaka factor, Klf4 induces the expression of genes that promote stem cell characteristics, such as NANOG, by suppressing p53 protein.

Figure 5: Flow chart of the process of reprogramming via Yamanaka factors. Image taken
from Kulcenty et al., 2015, Molecular mechanisms of induced pluripotency.
Since the Nobel-prize-winning discovery, the induction of pluripotency via OSKM, along with other transcription factors, such as Nanog and Lin28, has been replicated in many cell lines, and even in human cells (Stadtfeld and Hochedlinger, 2010).
Small chemical molecules that can mimic the effect of those transcription factors have also been deployed into the battlefield. In 2008, a proof-of-concept study was published, where a histone-deacetylase inhibitor, valproic acid could not only induce pluripotency but improve reprogramming efficiency by 100 folds (Huangfu et al., 2008). Subsequently, in 2013, researchers from Beijing University also showed that somatic cells could be reprogrammed into stem cells by using a combination of seven small molecules (Hou et al., 2013). Up till now, there have been multiple published articles proving that pluripotency can be induced by chemical molecules, with the most recent one being in July 2023 (Kim, Jeong and Choi, 2020, Guan et al., 2022, Yang et al., 2023).
Implications in medical research
The ability to produce pluripotent stem cells from almost any cell lineage has large implications in stem cell therapy. Prior to Yamanaka’s article about inducing pluripotency in human somatic cells, PSCs could only be obtained from pre-implantation embryos, called embryonic stem cells (ESCs) which although opened new avenues for biomedical research, was packaged along with multiple challenges (Hasegawa, Pomeroy and Pera, 2010). Ethical limitations, due to the requirement to destroy human embryos, has made its way into many countries’ policy restricting the use of ESCs; immunological rejection of the cells differentiated from ESCs after stem cell transplantation for a donor also limited its practicability (similar to how organ transplantation needs a matching donor and receiver) (Taylor, Bolton and Bradley, 2011). Given that iPSCs can be produced from somatic cells of the receiver, it circumvents those two challenges, as no embryo would need to be destroyed and no immune reaction would be launched against the implanted cells.
The basic paradigm of stem cell therapy is that PSCs are first differentiated into the desired cell types of interest, then transplanted as cell suspensions or more complex tissue constructs into patients. The differentiation step is vital as proliferating, undifferentiated PSCs can form tumors called teratomas if injected directly due to their highly proliferative nature and broad differentiation potential (Abad et al., 2013).

Figure 6: Basic paradigm of iPSC-based stem cell therapy. Image taken from BPS
Bioscience.
Because of immortality and multi-lineage differentiation potential, iPSCs are equally suitable for all the potential biomedical applications of ESCs. In cases where iPSCs are derived from patients with genetic defects, it may be possible to combine them with gene editing technology such as CRISPR-Cas9 to correct the genetic abnormalities prior to transplantation (Giacalone et al., 2018).
So far since the discovery of iPSCs, three autologous transplants of iPSCs in humans have been conducted. None of these patients have suffered adverse serious effects despite the lack of immunosuppression.
The first autologous iPSCs-derived cell type human transplantation were retinal pigment epithelial cells (RPE). This transplant was performed to treat age-related macular degeneration (AMD), a chronic disease characterized by the degeneration of RPE cells and the leading cause of vision loss among individuals over age 60. In this study, an iPSC-RPE sheet measuring 1.3 × 3 mm was transplanted under the fovea of the right eye of a 77-year-old female patient with neovascular AMD. In the 4-year follow-up, the patient reported not experiencing any adverse effects, while also maintaining stable vision despite a consistent decrease in vision prior to the study (Mandai et al., 2017).
The second transplantation involves the transplantation of iPSC-derived midbrain dopaminergic (mDA) progenitor cells (mDAPs) into the putamen of a 69-year-old man with a 10-year history of Parkinson’s disease. Parkinson’s disease is a neurodegenerative disease, characterized by the loss of dopaminergic neurons, and can lead to motor symptoms such as tremors and slow movements. Usually when symptoms are obvious enough to lead to a diagnosis, ~60% of mDA neurons have degenerated, therefore regeneration of those neurons is crucial for improving the quality of life of a patient with Parkinson’s disease (Engelender and Isacson, 2017). After transplantation, the patient's clinical measurements were taken at 1, 3, 6, 9, and 12 months post-transplantation and then at 6-month intervals. The patient's self-reported Unified Parkinson’s Disease rating scale (UPDRS) score (the lower the better), decreased from 60 at the time of implantation to 2 in the 24-month follow-up. To date, no serious adverse events have been recorded (Schweitzer et al., 2020).
The third transplantation was in a patient who suffered from aplastic anemia, a rare disease characterized by thrombocytopenia (low platelet count), and further accentuated by being refractory towards platelet transfusion treatment. The patient received three doses of autologous iPSC-derived platelets: 1 × 10^10 platelets, 3 × 10^10 platelets 3 months later, and then 1 × 10^11 platelets 5 months after the second dose. At the 1-year follow-up, the patient did not report any adverse events, but the investigators failed to observe any rise in platelet count, though larger platelets were detected via flow cytometry (Sugimoto et al., 2022).
Besides these in-human trials, there have also been pre-clinical trials investigating iPSCs-derived cell-type transplantation in animal models of diseases. There have been studies done to treat epidermolysis bullosa (Jacków et al., 2019, Sebastiano et al., 2014), muscular dystrophies (Sun et al., 2020), retinal degenerative blindness (Wiley et al., 2016), and perhaps the most promising sickle cell anemia, as it completely rescued the disease phenotype when transplanted mice with sickle cell anemia received hematopoietic progenitor cells without genetic defects (Hanna et al., 2007). It is evident that iPSCs harbor plenty of potential for stem cell therapy due to the ability to mass produce and the lack of host rejection. However, more studies need to be done on more subjects to identify standardized procedural guidelines, long-term safety profiles, and more diseases that can be treated.
Other than stem cell therapy, iPSCs can also be used for disease modeling and drug development (Tang et al., 2015). Since iPSCs are self-renewing and pluripotent, they can essentially become an unlimited source of patient-derived cells that can become any desired cell type. This is important as many other patient-derived cells tend to stop growing after a while in laboratory cultures. Thus far, iPSCs have been generated for many human diseases, including common ones such as Down syndrome (Park et al., 2008) and polycystic kidney disease (Freedman et al., 2013). The difference between disease-specific iPSCs and healthy iPSCs may therefore provide insight into the pathophysiology of diseases (Grskovic et al.,
2011).
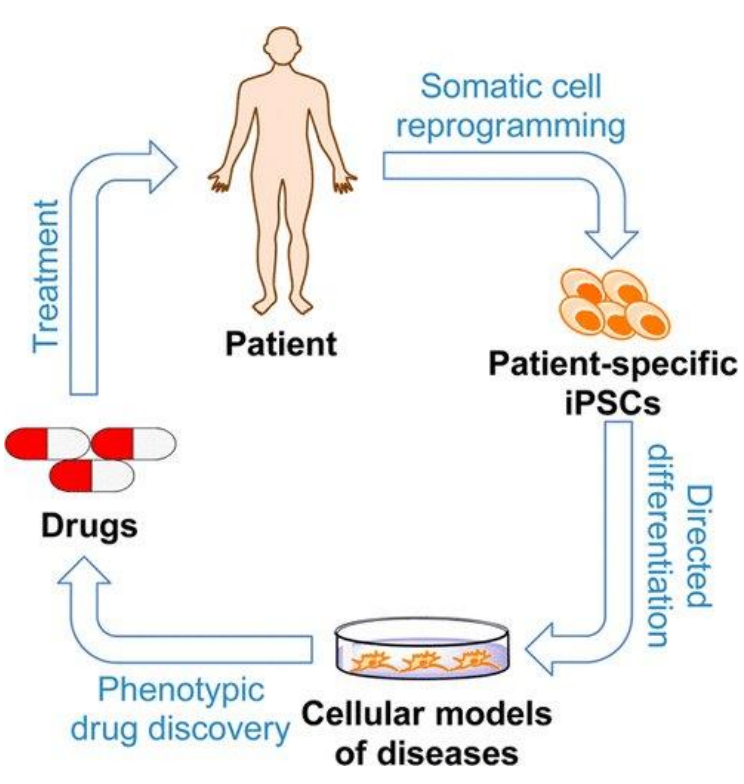
Figure 7: Usage of iPSCs for disease modeling and drug discovery. Image taken from Tang
et al., 2015, American Chemical Society.
Another potential application of iPSCs is to help satisfy the need for O-type blood. Many studies have shown the ability of iPSCs to differentiate into erythrocytes, but their enucleating capacity remains very low, though it can be increased through co-culture with different molecules (Satchwell, 2022).
Another recent research did not utilize OSKM but used a cocktail of small molecules and small interfering RNA to activate downstream pathways of reprogramming factors such as Myc. The cocktail was successful in reprogramming adult cochlea for hair cell-like cells regeneration in wild-type (WT) mice with hair cell loss in vivo. This could potentially be a treatment for hearing recovery in patients with permanent hearing loss (Quan et al., 2023).
Rejuvenation of aging
When talking about aging though, we wouldn’t want all the somatic cells in our body to undergo complete dedifferentiation, as you'd just end up with non-functional stem cells which could potentially divide and differentiate uncontrollably to form teratomas. In fact, iPSCs are very prone to forming teratomas if directly injected into a host, which is why researchers differentiate the iPSCs into cells of interest first before transplantation (Gutierrez-Aranda et al., 2010). Imagine reprogramming a beating heart into stem cells, you'd die relatively fast.
Researchers have proposed that by manipulating the duration of reprogramming, composition of reprogramming cocktails used, and dose of expression, they could reprogram cells just far enough to regain their youthful function, but not too far that they lose their identity and become pluripotent stem cells. This process is known as partial reprogramming, or more accurately as reprogramming-induced rejuvenation (RIR).
In 2013, Kurian and colleagues showed that reprogramming proceeds in a stepwise manner, which allowed for the induction of partial reprogramming without the complete loss of cellular identity by short exposure to OSKM (Kurian et al., 2013). In the same year, Tanabe and colleagues showed that expression of OSKM for 7 days resulted in a partially reprogrammed state, whereby high expression of pluripotency genes was observed, but those cells quickly regained somatic identity after cessation of OSKM expression (Tanabe et al., 2013).
In 2016, the first landmark study to show that RIR could rejuvenate cells in-vivo was published. The researchers used mice that were homozygous in the mutation that caused them to produce defective lamin A protein, in order to model human Hutchinson-Gilford progeria syndrome. These mice experience accelerated aging. The mice were also genetically modified via breeding to express OSKM upon administration of the inert antibiotic doxycycline. The researchers used a cyclic induction protocol of 2 days of doxycycline administration, followed by 5 days of doxycycline withdrawal, repeated over the lifespan of the mice. This protocol increased the median and maximum lifespan of mice and ameliorated
many age-associated features, such as a reduction in spine curvature, gross improvement in the appearance of the gastrointestinal tract, increased epidermal and dermal thickness, decreased keratinization of the skin, and many more. To more accurately resemble physiological aging, the researchers repeated the weekly cycle 3 times in 12-month-old wild-type mice (equivalent to around a 40-year-old human). They showed that the protocol significantly enhanced the mouse’s pancreas and muscle regenerative capacity (Ocampo et al., 2016). This could be beneficial in the future as diabetes and sarcopenia are both common diseases of the old (Jaul and Barron, 2017). One thing to note is the use of transgenic animals can have up to 50-fold higher efficiency and accelerated kinetics of reprogramming (Wernig et al., 2008), which may explain why only 2 days of OSKM induction was needed to achieve partial reprogramming as compared to those shown by other labs (Olova et al., 2018).

Figure 8: Graphical abstract of Ocampo et al., 2016. Image taken from Ocampo et al.,
Cell.
Similarly, another study performed transient expression of nuclear reprogramming factors, including OSKM, Lin28, and Nanog, mediated by the expression of mRNAs, in multiple cell lines in vitro. The study showed that the transient reprogramming promoted a rapid and broad amelioration of cellular aging, including resetting of the epigenetic clock, reduction of the inflammatory profile in chondrocytes, and restoration of youthful regenerative response to aged, human muscle stem cells, in each case without compromising cellular identity (Sarkar et al., 2020).
In 2022, Alle and colleagues further showed that a 2.5-week continuous medium dose of OSKM induction in young (2 months old) heterozygous progeroid mice increased lifespan as much as the lifelong cyclic (2 days on, 5 days off) high dose of OSKM induction. The single early-life treatment also increased the lean mass-to-fat ratio, provided protection against age-related skin thinning and loss of elasticity, preserved the function of the lungs, spleens, and kidneys, and improved bone mass (Alle et al., 2022).

Figure 9: Experimental procedure of Alle et al., 2022. Image taken from Alle et al., 2022,
National Library of Medicine.
In contrast to other studies that used multiple cycles of OSKM induction, Chondronasiou and colleagues showed that only one one-week in-vivo induction of OSKM in middle-aged WT mice was able to cause epigenetic, transcriptomic, and metabolomic changes toward a younger configuration in multiple tissues and in the serum. However, they also showed that some of the rejuvenation effects are unstable and disappear two weeks after the OSKM withdrawal (Chondronasiou et al., 2022).
A new preprint has recently shone light on the potential of OSK to reverse aging in WT mice specifically, compared to Ocampo and colleagues' progeroid mice. These researchers used an adeno-associated virus (AAV) to modify WT mice to express OSK in the presence of doxycycline on an alternate weekly on/off cycle. Their justification for the omission of c-Myc is that it is a proto-oncogene. The strengths of this study include their target population, which are old mice (124 weeks, 77-years-old human equivalent), as young populations would usually not be targets of age-reversal therapy. They observed a significant 109% increase in the median remaining lifespan and a significant reduction in frailty in response to OSK expression (Macip et al., 2023). This preprint however is not yet peer-reviewed.

Figure 10: Macip et al., 2023 study results. Image taken from Macip et al., 2023, bioRxiv.
Besides systemic whole-body reprogramming, researchers have also opted for organ-specific rejuvenation as it is easier to control the dosing and duration of reprogramming when not involving many different cells and organs.
In 2020, a groundbreaking article was released from Dr. David Sinclair’s lab showing that transient expression of OSK factors for 10-18 months in mouse retinal ganglion cells restores youthful DNA methylation patterns and transcriptomes, promotes axon regeneration after injury, and reverses vision loss in a mouse model of glaucoma and in aged mice. Not only that, the omission of c-Myc due to its oncogenic properties seems to lead to no tumor formation in treated animals (Lu et al., 2020). Recently, Dr. David Sinclair has led another team of researchers to show that the expression of OSK was able to restore parameters of visual pathway function in a non-human primate model of non-arteritic anterior ischemic optic neuropathy (NAION) (Ksander et al., 2023). The full paper has not yet been published, so any conclusions drawn should be conservative.
Post-mitotic terminally differentiated cells such as cardiomyocytes and neurons generally possess low regenerative capability. This is because those cells tend to stop dividing after reaching a certain age. Hence, regeneration of organs such as the heart is extremely difficult (Bergmann et al., 2015), yet important due to the implications of a heart attack. Transient in-vivo expression of cardiomyocyte-specific OSKM seemed to allow mitosis-refractory adult cardiomyocytes to reenter a mitotic-competent state. The researchers further demonstrate that the transient expression of cardiomyocytes-specific OSKM for 6 days in-vivo could allow adult mice hearts that have undergone myocardial infarction (MI) to regenerate, and what's more in the absence of tumor formation. This effect was only significant in mice treated 6 days pre-MI, or 1 day post-MI, not 6 days post-MI, indicating a time-dependent response. However, controlling the duration of OSKM expression is vital as the expression of the factors for 12 days results in excess dedifferentiation and reduced heart function (Chen et al., 2021).

Figure 11: Comparison between the healing of the neonatal heart and adult heart after an
MI. Image taken from ResearchGate.
Some teams of researchers sought to circumvent the post-mitotic nature of adult cardiomyocytes by reprogramming fibroblasts within cardiac scar tissue back into cardiomyocytes through miRNA (Baksh and Hodgkinson, 2022), or combinations of transcription factors (Song et al., 2012). However, improvements in cardiac function were modest. A team of researchers proposed that phenotypical changes could happen during the transition of neonatal fibroblasts into adult fibroblasts that induce resistance to reprogramming in adult fibroblasts. They found that the transition from the neonatal to adult cardiac fibroblast phenotype was dependent on the transcription factor Epas1. When adult cardiac fibroblasts were reverted to their neonatal phenotype via Epas1 knockdown, reprogramming efficacy improved, as well as enhanced functional improvements associated with reprogramming in infarcted adult mice (Sun et al., 2023). This study suggests a synergistic effect of other genetic engineering procedures with reprogramming protocols.
One study showed that in vivo partial reprogramming in skeletal muscles created overlapping gene expressions with those induced by late-life exercise, suggesting an alternative for frail elderly that have a hard time exercising (Jones et al., 2023).
For cognition, a study in 2020 showed that in-vivo reprogramming in the central nervous system elevated the levels of migrating cells containing the neurogenic markers doublecortin and calretinin, and the levels of the NMDA receptor subunit GluN2B. They suggested this could increase the survival of newborn dentate gyrus neurons, as well as increase synaptic plasticity in mature neurons. In fact, the reprogramming successfully improved memory in old mice (Rodríguez-Matellán et al., 2020).
Though most studies showed no loss of cellular identity, one recent study showed that partial reprogramming restores youthful gene expression by transiently suppressing cell identity. What’s different is that the study used single-cell sequencing to identify changes in cell identity which is more refined and accurate than commonly used bulk-sequencing methods (Yu et al., 2020). This suggests that even partial reprogramming could be oncogenic in a detectable subpopulation of cells. However, the study showed that the loss of cellular identity could be decoupled from the rejuvenation of aging, as different combinations of the OSKM factors could reduce age scores, and identity scores in a way where both variables are not correlated (Roux et al., 2022).

Figure 12: Results of Roux et al., 2022 showing age score and mesenchymal identity are
uncoupled during reprogramming with different combinations of OSKM. Image taken
from Roux et al., 2022, Cell Systems.
Another recent preprint also showed that the genes involved in rejuvenation are different from genes involved in the gain of pluripotency. Knowing this enables future research to identify compounds or factors that can activate rejuvenation genes without activating pluripotency genes (Kriukov et al., 2022).
Practical applications
Besides its current use in developing stem cells for disease modeling and transplantation, its application may extend into non stem cell based ones. If proven effective in humans, the medical field may start using epigenetic reprogramming for prevention of age-related disease in at-risk populations. More targeted applications can involve using epigenetic reprogramming to repair a certain organ or tissue from damage. What's best is to be able to use such technology to completely prevent and eradicate aging, even in young populations to keep everyone healthy, active, and able to enjoy their life.
Setting the expectations straight
Although it may sound like all sunshine and rainbows, there are still problems plaguing epigenetic programming that hinders its translation into human clinical trials.
For stem cell therapy, the low efficiency of reprogramming is a major roadblock. Yamanaka’s original mouse study only had an efficiency of 0.1%-0.01% (Takahashi and Yamanaka, 2006). However, recent work found a path for efficient reprogramming: depletion of Mbd3, a subunit of the nucleosome remodeling and deacetylation (NuRD) complex, improves reprogramming efficiency to nearly 100% within 7 days (Rais et al., 2013), and can even improve reprogramming efficiency by 10-fold in the absence of Sox2 or c-Myc (Luo et al., 2013).
Tumorigenicity of iPSCs-derived cells can also be a problem, especially if they were reprogrammed through a viral vector. This is because the virus could potentially integrate into the genome, causing unwanted expression of the reprogramming factors and mutations (Bayart and Cohen-Haguenauer, 2013), potentially leading to dedifferentiation and hence teratomas when transplanted. Furthermore, there seems to be a tradeoff between reprogramming efficiency and tumor generation, as inactivation or deletion of the tumor suppressor p53, which is a key regulator of cancer, significantly increases reprogramming efficiency (Marión et al., 2009).

Figure 13: Viral integration can activate oncogenes, as well as deactivate tumor suppressor
genes. Image taken from Wiley Online Library.
Perhaps the most notable problem for RIR is how will a human express the reprogramming factors. Most animal studies use viral vectors to introduce the reprogramming factors into somatic cells or use transgenic mice that express the reprogramming factors, both of which are doxycycline-inducible. The problem is that creating transgenic humans is unethical and that viral vectors may integrate into the human genome, causing unwanted mutations. Even non-integrating vectors such as AAV or adenoviruses have an albeit much lower chance of integrating into the host (Deyle and Russell, 2009). Alternative gene delivery methods such as episomal vectors, transient DNA transfection, transposon-derived excisable vectors, and protein transduction can circumvent this problem, but they have much lower efficiency of reprogramming (Bayart and Cohen-Haguenauer, 2013). One novel method that does not integrate and have high reprogramming efficiency is an mRNA-based method, though this method can cause foreign mRNA cytotoxicity (Drews et al., 2012).
Besides, identifying the duration of expression of reprogramming factors in humans should also be looked into. This is because the risks of “over-reprogramming” are very high, as it could counterintuitively cause teratomas. Furthermore, it is possible for different populations of humans to require a different duration of expression. From mice studies, the duration of reprogramming varies between mouse models, with progeroid mice needing a shorter induction period, compared to WT mice, evidenced by the contrast in the presence and sustainability of OSKM induction’s effect between Alle and colleagues, Browder and colleagues, and Chondronasiou and colleagues (Alle et al., 2022, Browder et al., 2022, Chondronasiou et al., 2022). Age of initiation and delivery method probably influences the duration of reprogramming too, as one recent study showed that a one-month reprogramming period in middle-aged wild-type mice via AAV vector did not rejuvenate tissues, while a seven-month and 10 months reprogramming period did (Browder et al., 2022).
Systemic reprogramming seems to also be unequal across different organs evidenced by Chondronasiou and colleagues' experiment showing that rejuvenation was higher in the pancreas than in the liver and spleen (Chondronasiou et al., 2022). Hence, the cocktail of reprogramming factors may need to be tailored to specific cell types, to ensure correct amounts are distributed to the cells.
Other than that, the decoupling between dedifferentiation and rejuvenation also needs to be more deeply researched. Ideally, reprogramming factors or novel compounds that will induce rejuvenation genes while keeping dedifferentiation genes untouched should be identified and studied more.
Another factor to keep in mind is that all studies investigating RIR thus far have been in animal models, which while informative, is not necessarily translatable to humans. Given the highly toxic effects of teratoma formation if the duration of reprogramming is not controlled properly, I personally feel that much more work has to be done to allow human RIR trials.
Lastly, the overemphasis of cellular rejuvenation may distract our attention from other problems. Some of these problems may also contribute to the signs of aging and disease. For example, DNA mutations have been proposed to be a factor of aging (Schumacher et al., 2021). In fact DNA mutations are more prevalent in older organisms. Although most mutations appear to be harmless, harmful mutations do occur such as in the case of cancer. Epigenetic reprogramming cannot directly correct DNA mutations, as it only works on the principle of reversible modifiers of DNA expression. Regardless, our body's ability to correct DNA mutation decreases as we age, via downregulation of key DNA repair enzymes (Gorbunova et al., 2007). Hence, theoretically if we are able to reverse the effects of aging on cells and upregulate DNA repair enzymes, we may be able to indirectly correct present gene mutations.
Another possibly overlooked factor is the extracellular matrix (ECM), the environment for many cells, a structural support, and an influencer of many cellular processes. It is well-known that the proteins constituting the ECM change with age, which disrupt many different aspects of homeostasis and healthy function (Birch, 2018). Epigenetic reprogramming works on the cellular level, which does not directly affect the ECM, though reprogrammed fibroblasts may be able to secrete more collagen and elastin, while simultaneously shifting their secreted cytokines from a pro-inflammatory profile to an anti-inflammatory one (Sarkar et al., 2020). Indirectly, the ECM can be altered in a positive manner. Hence, more research should be done to identify whether epigenetic reprogramming can directly affect the other hallmarks of aging, so as to form a unifying theory of aging.

Figure 14: Hallmarks of aging. Image taken from Cell.
Takeaways
In conclusion, epigenetic reprogramming is perhaps one of the sharpest in the toolbox of health and longevity. Through harnessing the power of epigenetic modifications, we stand on the brink of unlocking new therapeutic avenues, paving the way for personalized medicine and targeted interventions that could transform the landscape of healthcare. However, the lack of thorough understanding of the sharpest in the toolbox, it is very dangerous if proper guidelines and framework are not used. More research has to be done to establish proper dosage, duration, and other logistical and physiological factors to ensure efficacy and safety. Human trials should also be done in order to properly verify the validity of epigenetic reprogramming. Let us, therefore, exercise patience and anticipation, eagerly awaiting the potential miracles that epigenetic reprogramming may bestow upon us. Adiós.
(5499 words)
Reference list
Abad, M., Mosteiro, L., Pantoja, C., Cañamero, M., Rayon, T., Ors, I., Graña, O., Megías, D., Domínguez, O., Martínez, D., Manzanares, M., Ortega, S. and Serrano, M. (2013). Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature, 502(7471), pp.340–345. doi:https://doi.org/10.1038/nature12586.
Al, N.M., Simpson, B. and Ishwarlal Jialal (2019). Genetics, Epigenetic Mechanism. [online] Nih.gov. Available at: https://www.ncbi.nlm.nih.gov/books/NBK532999/.
Alle, Q., Le Borgne, E., Bensadoun, P., Lemey, C., Béchir, N., Gabanou, M., Estermann, F., Bertrand‐Gaday, C., Pessemesse, L., Toupet, K., Desprat, R., Vialaret, J., Hirtz, C., Noël, D., Jorgensen, C., Casas, F., Milhavet, O. and Lemaitre, J. (2022). A single short reprogramming early in life initiates and propagates an epigenetically related mechanism improving fitness and promoting an increased healthy lifespan. Aging Cell, 21(11). doi:https://doi.org/10.1111/acel.13714.
Baksh, S.S. and Hodgkinson, C.P. (2022). Conservation of miR combo based direct cardiac reprogramming. Biochemistry and Biophysics Reports, [online] 31, p.101310. doi:https://doi.org/10.1016/j.bbrep.2022.101310.
Bannister, A.J. and Kouzarides, T. (2011). Regulation of Chromatin by Histone Modifications. Cell Research, [online] 21(3), pp.381–395. doi:https://doi.org/10.1038/cr.2011.22.
Bayart, E. and Cohen-Haguenauer, O. (2013). Technological Overview of iPS Induction from Human Adult Somatic Cells. Current Gene Therapy, 13(2), pp.73–92. doi:https://doi.org/10.2174/1566523211313020002.
Bergmann, O., Zdunek, S., Felker, A., Salehpour, M., Alkass, K., Bernard, S., Sjostrom, Staffan L., Szewczykowska, M., Jackowska, T., dos Remedios, C., Malm, T., Andrä, M., Jashari, R., Nyengaard, Jens R., Possnert, G., Jovinge, S., Druid, H. and Frisén, J. (2015). Dynamics of Cell Generation and Turnover in the Human Heart. Cell, [online] 161(7), pp.1566–1575. doi:https://doi.org/10.1016/j.cell.2015.05.026.
Birch, H.L. (2018). Extracellular Matrix and Ageing. Sub-Cellular Biochemistry, [online] 90, pp.169–190. doi:https://doi.org/10.1007/978-981-13-2835-0_7.
Browder, K.C., Reddy, P., Yamamoto, M., Haghani, A., Guillen, I.G., Sahu, S., Wang, C., Luque, Y., Prieto, J., Shi, L., Shojima, K., Hishida, T., Lai, Z., Li, Q., Choudhury, F.K., Wong, W.R., Liang, Y., Sangaraju, D., Sandoval, W. and Esteban, C.R. (2022). In vivo partial reprogramming alters age-associated molecular changes during physiological aging in mice. Nature Aging, [online] 2(3), pp.243–253. doi:https://doi.org/10.1038/s43587-022-00183-2.
Buganim, Y., Faddah, Dina A., Cheng, Albert W., Itskovich, E., Markoulaki, S., Ganz, K., Klemm, Sandy L., van Oudenaarden, A. and Jaenisch, R. (2012). Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell, 150(6), pp.1209–1222. doi:https://doi.org/10.1016/j.cell.2012.08.023.
Chen, Y., Lüttmann, F.F., Schoger, E., Schöler, H.R., Zelarayán, L.C., Kim, K.-P., Haigh, J.J., Kim, J. and Braun, T. (2021). Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science, 373(6562), pp.1537–1540. doi:https://doi.org/10.1126/science.abg5159.
12. Chondronasiou, D., Gill, D., Mosteiro, L., Urdinguio, R.G., Berenguer‐Llergo, A.,
Aguilera, M., Durand, S., Aprahamian, F., Nirmalathasan, N., Abad, M.,
Martin‐Herranz, D.E., Stephan‐Otto Attolini, C., Prats, N., Kroemer, G., Fraga, M.F.,
Reik, W. and Serrano, M. (2022). Multi‐omic rejuvenation of naturally aged tissues
by a single cycle of transient reprogramming. Aging Cell, 21(3).
doi:https://doi.org/10.1111/acel.13578.
13. Deyle, D.R. and Russell, D.W. (2009). Adeno-associated virus vector integration.
Current opinion in molecular therapeutics, [online] 11(4), pp.442–447. Available at:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2929125/.
14. Douglas, P.M. and Dillin, A. (2010). Protein homeostasis and aging in
neurodegeneration. Journal of Cell Biology, 190(5), pp.719–729.
doi:https://doi.org/10.1083/jcb.201005144.
15. Drews, K., Tavernier, G., Demeester, J., Lehrach, H., De Smedt, S.C., Rejman, J. and
Adjaye, J. (2012). The cytotoxic and immunogenic hurdles associated with non-viral
mRNA-mediated reprogramming of human fibroblasts. Biomaterials, [online] 33(16),
pp.4059–4068. doi:https://doi.org/10.1016/j.biomaterials.2012.02.025.
16. Engelender, S. and Isacson, O. (2017). The Threshold Theory for Parkinson’s Disease.
Trends in Neurosciences, [online] 40(1), pp.4–14.
doi:https://doi.org/10.1016/j.tins.2016.10.008.
17. Freedman, B.S., Lam, A.Q., Sundsbak, J.L., R Iatrino, Su, X., Koon, S.J., Wu, M.,
Daheron, L., Harris, P.C., Zhou, J. and Bonventre, J.V. (2013). Reduced Ciliary
Polycystin-2 in Induced Pluripotent Stem Cells from Polycystic Kidney Disease Patients with PKD1 Mutations. Journal of The American Society of Nephrology,
24(10), pp.1571–1586. doi:https://doi.org/10.1681/asn.2012111089.
18. Giacalone, J.C., Sharma, T.P., Burnight, E.R., Fingert, J.F., Mullins, R.F., Stone, E.M.
and Tucker, B.A. (2018). CRISPR-Cas9-Based Genome Editing of Human Induced
Pluripotent Stem Cells. Current protocols in stem cell biology, [online] 44,
pp.5B.7.1–5B.7.22. doi:https://doi.org/10.1002/cpsc.46.
19. Gill, D., Parry, A., Santos, F., Okkenhaug, H., Todd, C.D., Hernando-Herraez, I.,
Stubbs, T.M., Milagre, I. and Reik, W. (2022). Multi-omic rejuvenation of human
cells by maturation phase transient reprogramming. eLife, 11.
doi:https://doi.org/10.7554/elife.71624.
20. Gorbunova, V., Seluanov, A., Mao, Z. and Hine, C. (2007). Changes in DNA repair
during aging. Nucleic Acids Research, 35(22), pp.7466–7474.
doi:https://doi.org/10.1093/nar/gkm756.
21. Grskovic, M., Javaherian, A., Strulovici, B. and Daley, G.Q. (2011). Induced
pluripotent stem cells — opportunities for disease modelling and drug discovery.
Nature Reviews Drug Discovery, 10, pp.915–929.
doi:https://doi.org/10.1038/nrd3577.
22. Guan, J., Wang, G., Wang, J., Zhang, Z., Fu, Y., Cheng, L., Meng, G., Lyu, Y., Zhu, J.,
Li, Y., Wang, Y., Liuyang, S., Liu, B., Yang, Z., He, H., Zhong, X., Chen, Q., Zhang,
X., Sun, S. and Lai, W. (2022). Chemical reprogramming of human somatic cells to
pluripotent stem cells. Nature, [online] 605, pp.1–7.
doi:https://doi.org/10.1038/s41586-022-04593-5.
23. Gurdon, J.B. (1962). Adult frogs derived from the nuclei of single somatic cells.
Developmental Biology, 4(2), pp.256–273.
doi:https://doi.org/10.1016/0012-1606(62)90043-x.
24. Gutierrez-Aranda, I., Ramos-Mejia, V., Bueno, C., Munoz-Lopez, M., Real, Pedro.J.,
Mácia, A., Sanchez, L., Ligero, G., Garcia-Parez, J.L. and Menendez, P. (2010).
Human Induced Pluripotent Stem Cells Develop Teratoma More Efficiently and
Faster Than Human Embryonic Stem Cells Regardless the Site of Injection. STEM
CELLS, 28(9), pp.1568–1570. doi:https://doi.org/10.1002/stem.471.
25. Hanna, J., Wernig, M., Markoulaki, S., Sun, C.-W. ., Meissner, A., Cassady, J.P.,
Beard, C., Brambrink, T., Wu, L.-C. ., Townes, T.M. and Jaenisch, R. (2007).
Treatment of Sickle Cell Anemia Mouse Model with iPS Cells Generated from Autologous Skin. Science, [online] 318(5858), pp.1920–1923. doi:https://doi.org/10.1126/science.1152092.
26. Hasegawa, K., Pomeroy, J.E. and Pera, M.F. (2010). Current Technology for the
Derivation of Pluripotent Stem Cell Lines from Human Embryos. Cell Stem Cell,
6(6), pp.521–531. doi:https://doi.org/10.1016/j.stem.2010.05.010.
27. Hou, P., Li, Y., Zhang, X., Liu, C., Guan, J., Li, H., Zhao, T., Ye, J., Yang, W., Liu, K.,
Ge, J., Xu, J., Zhang, Q., Zhao, Y. and Deng, H. (2013). Pluripotent Stem Cells
Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science, [online]
341(6146), pp.651–654. doi:https://doi.org/10.1126/science.1239278.
28. Hou, Y., Dan, X., Babbar, M., Wei, Y., Hasselbalch, S.G., Croteau, D.L. and Bohr,
V.A. (2019). Ageing as a risk factor for neurodegenerative disease. Nature Reviews.
Neurology, 15(10), pp.565–581. doi:https://doi.org/10.1038/s41582-019-0244-7.
29. Huangfu, D., Maehr, R., Guo, W., Eijkelenboom, A., Snitow, M., Chen, A.E. and
Melton, D.A. (2008). Induction of pluripotent stem cells by defined factors is greatly
improved by small-molecule compounds. Nature Biotechnology, [online] 26(7),
pp.795–797. doi:https://doi.org/10.1038/nbt1418.
30. Hutchings, M.I., Truman, A.W. and Wilkinson, B. (2019). Antibiotics: past, Present
and Future. Current Opinion in Microbiology, [online] 51(1), pp.72–80.
doi:https://doi.org/10.1016/j.mib.2019.10.008.
31. Jacków, J., Guo, Z., Hansen, C., Abaci, H.E., Doucet, Y.S., Shin, J.U., Hayashi, R.,
DeLorenzo, D., Kabata, Y., Shinkuma, S., Salas-Alanis, J.C. and Christiano, A.M.
(2019). CRISPR/Cas9-based targeted genome editing for correction of recessive
dystrophic epidermolysis bullosa using iPS cells. Proceedings of the National
Academy of Sciences, [online] 116(52), pp.26846–26852.
doi:https://doi.org/10.1073/pnas.1907081116.
32. Jaul, E. and Barron, J. (2017). Age-Related Diseases and Clinical and Public Health
Implications for the 85 Years Old and Over Population. Frontiers in Public Health,
5(335). doi:https://doi.org/10.3389/fpubh.2017.00335.
33. Jones, R.G., Dimet‐Wiley, A., Haghani, A., da Silva, F.M., Brightwell, C.R., Lim, S.,
Khadgi, S., Wen, Y., Dungan, C.M., Brooke, R.T., Greene, N.P., Peterson, C.A.,
McCarthy, J.J., Horvath, S., Watowich, S.J., Fry, C.S. and Murach, K.A. (2023). A
molecular signature defining exercise adaptation with ageing and in vivo partial
reprogramming in skeletal muscle. The Journal of Physiology, 601(4), pp.763–782.
doi:https://doi.org/10.1113/jp283836.
34. Kim, Y., Jeong, J. and Choi, D. (2020). Small-molecule-mediated reprogramming: a
silver lining for regenerative medicine. Experimental & Molecular Medicine, 52(2),
pp.213–226. doi:https://doi.org/10.1038/s12276-020-0383-3.
35. Kriukov, D., Khrameeva, E.E., Gladyshev, V.N., Dmitriev, S.E. and Tyshkovskiy, A.
(2022). Longevity and rejuvenation effects of cell reprogramming are decoupled from
loss of somatic identity. bioRxiv. doi:https://doi.org/10.1101/2022.12.12.520058.
36. Ksander, B., Shah, M., Krasniqi, D., Gregory-Ksander, M.S., Rosenzweig-Lipson, S.,
Kasia Broniowska, Wathier, M., Mannick, J., Cermak, J., Karg, M., Shintaro
Shirahama, Nasrin Refaian, Lu, Y., Lawrence, M., Rizzo, J.F. and Sinclair, D. (2023).
Epigenetic reprogramming- A novel gene therapy that restores vision loss in a
nonhuman primate model of NAION. Investigative Ophthalmology & Visual Science,
[online] 64(8), pp.474–474. Available at:
https://iovs.arvojournals.org/article.aspx?articleid=2785785
37. Kulcenty, K., Wróblewska, J., Mazurek, S., Liszewska, E. and Jaworski, J. (2015).
Review Molecular mechanisms of induced pluripotency. Współczesna Onkologia, 1A,
pp.22–29. doi:https://doi.org/10.5114/wo.2014.47134.
38. Kurian, L., Sancho-Martinez, I., Nivet, E., Aguirre, A., Moon, K., Pendaries, C.,
Volle-Challier, C., Bono, F., Herbert, J.-M., Pulecio, J., Xia, Y., Li, M., Montserrat,
N., Ruiz, S., Dubova, I., Rodriguez, C., Denli, A.M., Boscolo, F.S., Thiagarajan, R.D.
and Gage, F.H. (2013). Conversion of human fibroblasts to angioblast-like progenitor
cells. Nature Methods, [online] 10(1), pp.77–83.
doi:https://doi.org/10.1038/nmeth.2255.
39. Li, B., Carey, M. and Workman, J.L. (2007). The Role of Chromatin during
Transcription. Cell, 128(4), pp.707–719.
doi:https://doi.org/10.1016/j.cell.2007.01.015.
40. Lu, Y., Brommer, B., Tian, X., Krishnan, A., Meer, M., Wang, C., Vera, D.L., Zeng,
Q., Yu, D., Bonkowski, M.S., Yang, J.-H., Zhou, S., Hoffmann, E.M., Karg, M.M.,
Schultz, M.B., Kane, A.E., Davidsohn, N., Korobkina, E., Chwalek, K. and Rajman,
L.A. (2020). Reprogramming to recover youthful epigenetic information and restore
vision. Nature, [online] 588(7836), pp.124–129.
doi:https://doi.org/10.1038/s41586-020-2975-4.
41. Luo, M., Ling, T., Xie, W., Sun, H., Zhou, Y., Zhu, Q., Shen, M., Zong, L., Lyu, G.,
Zhao, Y., Ye, T., Gu, J., Tao, W., Lu, Z. and Grummt, I. (2013). NuRD Blocks Reprogramming of Mouse Somatic Cells into Pluripotent Stem Cells. STEM CELLS, 31(7), pp.1278–1286. doi:https://doi.org/10.1002/stem.1374.
42. Macip, C.C., Hasan, R., Hoznek, V., Kim, J., Metzger, L.E., Sethna, S. and
Davidsohn, N. (2023). Gene Therapy Mediated Partial Reprogramming Extends
Lifespan and Reverses Age-Related Changes in Aged Mice. bioRXiV.
doi:https://doi.org/10.1101/2023.01.04.522507.
43. Mah, N., Wang, Y., Liao, M.-C., Alessandro Prigione, Justyna Jozefczuk, Lichtner, B.,
Wolfrum, K., Haltmeier, M., Flöttmann, M., Schaefer, M., Hahn, A.J., Mrowka, R.,
Klipp, E., Andrade-Navarro, M.A. and Adjaye, J. (2011). Molecular Insights into
Reprogramming-Initiation Events Mediated by the OSKM Gene Regulatory Network.
PLOS One, 6(8), pp.e24351–e24351.
doi:https://doi.org/10.1371/journal.pone.0024351.
44. Mandai, M., Watanabe, A., Kurimoto, Y., Hirami, Y., Morinaga, C., Daimon, T.,
Fujihara, M., Akimaru, H., Sakai, N., Shibata, Y., Terada, M., Nomiya, Y., Tanishima,
S., Nakamura, M., Kamao, H., Sugita, S., Onishi, A., Ito, T., Fujita, K. and
Kawamata, S. (2017). Autologous Induced Stem-Cell–Derived Retinal Cells for
Macular Degeneration. New England Journal of Medicine, 376(11), pp.1038–1046.
doi:https://doi.org/10.1056/nejmoa1608368.
45. Marión, R.M., Strati, K., Li, H., Murga, M., Blanco, R., Ortega, S.,
Fernandez-Capetillo, O., Serrano, M. and Blasco, M.A. (2009). A p53-mediated DNA
damage response limits reprogramming to ensure iPS cell genomic integrity. Nature,
[online] 460(7259), pp.1149–1153. doi:https://doi.org/10.1038/nature08287.
46. Moore, L.D., Le, T. and Fan, G. (2012). DNA Methylation and Its Basic Function.
Neuropsychopharmacology, [online] 38(1), pp.23–38.
doi:https://doi.org/10.1038/npp.2012.112.
47. North, B.J. and Sinclair, D.A. (2012). The Intersection Between Aging and
Cardiovascular Disease. Circulation Research, [online] 110(8), pp.1097–1108.
doi:https://doi.org/10.1161/circresaha.111.246876.
48. Ocampo, A., Reddy, P., Martinez-Redondo, P., Platero-Luengo, A., Hatanaka, F.,
Hishida, T., Li, M., Lam, D., Kurita, M., Beyret, E., Araoka, T., Vazquez-Ferrer, E.,
Donoso, D., Roman, J.L., Xu, J., Rodriguez Esteban, C., Nuñez, G., Nuñez Delicado,
E., Campistol, J.M. and Guillen, I. (2016). In Vivo Amelioration of Age-Associated
Hallmarks by Partial Reprogramming. Cell, [online] 167(7), pp.1719-1733.e12.
doi:https://doi.org/10.1016/j.cell.2016.11.052.
49. Olova, N., Simpson, D.J., Marioni, R.E. and Chandra, T. (2018). Partial
reprogramming induces a steady decline in epigenetic age before loss of somatic
identity. Aging Cell, [online] 18(1), p.e12877. doi:https://doi.org/10.1111/acel.12877.
50. Pal, S. and Tyler, J.K. (2016). Epigenetics and aging. Science Advances, [online] 2(7),
p.e1600584. doi:https://doi.org/10.1126/sciadv.1600584.
51. Park, I.-H., Arora, N., Huo, H., Maherali, N., Ahfeldt, T., Shimamura, A., Lensch,
M.W., Cowan, C., Hochedlinger, K. and Daley, G.Q. (2008). Disease-Specific
Induced Pluripotent Stem Cells. Cell, [online] 134(5), pp.877–886.
doi:https://doi.org/10.1016/j.cell.2008.07.041.
52. Polo, Jose M., Anderssen, E., Walsh, Ryan M., Schwarz, Benjamin A., Nefzger,
Christian M., Lim, S., Borkent, M., Apostolou, E., Alaei, S., Cloutier, J., Bar-Nur, O.,
Cheloufi, S., Stadtfeld, M., Figueroa, M., Robinton, D., Natesan, S., Melnick, A.,
Zhu, J., Ramaswamy, S. and Hochedlinger, K. (2012). A Molecular Roadmap of
Reprogramming Somatic Cells into iPS Cells. Cell, 151(7), pp.1617–1632.
doi:https://doi.org/10.1016/j.cell.2012.11.039.
53. Quan, Y.-Z., Wei, W., Volkan Ergin, Arun Prabhu Rameshbabu, Huang, M., Tian, C.,
Srinivas Vinod Saladi, Indzhykulian, A.A. and Chen, Z.-Y. (2023). Reprogramming
by drug-like molecules leads to regeneration of cochlear hair cell-like cells in adult
mice. Proceedings of the National Academy of Sciences of the United States of
America, 120(17). doi:https://doi.org/10.1073/pnas.2215253120.
54. Quina, A.S., Buschbeck, M. and Di Croce, L. (2006). Chromatin structure and
epigenetics. Biochemical Pharmacology, [online] 72(11), pp.1563–1569.
doi:https://doi.org/10.1016/j.bcp.2006.06.016.
55. Rais, Y., Zviran, A., Geula, S., Gafni, O., Chomsky, E., Viukov, S., Mansour, A.A.,
Caspi, I., Krupalnik, V., Zerbib, M., Maza, I., Mor, N., Baran, D., Weinberger, L.,
Jaitin, D.A., Lara-Astiaso, D., Blecher-Gonen, R., Shipony, Z., Mukamel, Z. and
Hagai, T. (2013). Deterministic direct reprogramming of somatic cells to pluripotency.
Nature, 502(7469), pp.65–70. doi:https://doi.org/10.1038/nature12587.
56. Rodríguez-Matellán, A., Alcazar, N., Hernández, F., Serrano, M. and Ávila, J. (2020).
In Vivo Reprogramming Ameliorates Aging Features in Dentate Gyrus Cells and
Improves Memory in Mice. Stem Cell Reports, 15(5), pp.1056–1066.
doi:https://doi.org/10.1016/j.stemcr.2020.09.010.
57. Roux, A.E., Zhang, C., Paw, J., Zavala-Solorio, J., Malahias, E., Vijay, T., Kolumam,
G., Kenyon, C. and Kimmel, J.C. (2022). Diverse partial reprogramming strategies restore youthful gene expression and transiently suppress cell identity. Cell Systems,
[online] 13(7), pp.574-587.e11. doi:https://doi.org/10.1016/j.cels.2022.05.002.
58. Rowland, B.A., Bernards, R. and Peeper, D.S. (2005). The KLF4 tumour suppressor
is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nature
Cell Biology, 7(11), pp.1074–1082. doi:https://doi.org/10.1038/ncb1314.
59. Sarkar, T.J., Quarta, M., Mukherjee, S., Colville, A., Paine, P., Doan, L., Tran, C.M.,
Chu, C.R., Horvath, S., Qi, L.S., Bhutani, N., Rando, T.A. and Sebastiano, V. (2020).
Transient non-integrative expression of nuclear reprogramming factors promotes
multifaceted amelioration of aging in human cells. Nature Communications, [online]
11(1). doi:https://doi.org/10.1038/s41467-020-15174-3.
60. Satchwell, T.J. (2022). Generation of red blood cells from stem cells: Achievements,
opportunities and perspectives for malaria research. Frontiers in Cellular and
Infection Microbiology, 12. doi:https://doi.org/10.3389/fcimb.2022.1039520.
61. Schumacher, B., Pothof, J., Vijg, J. and Hoeijmakers, J.H.J. (2021). The central role
of DNA damage in the ageing process. Nature, [online] 592(7856), pp.695–703.
doi:https://doi.org/10.1038/s41586-021-03307-7.
62. Schweitzer, J.S., Song, B., Herrington, T.M., Park, T.-Y., Lee, N., Ko, S., Jeon, J.,
Cha, Y., Kim, K., Li, Q., Henchcliffe, C., Kaplitt, M., Neff, C., Rapalino, O., Seo, H.,
Lee, I.-H., Kim, J., Kim, T., Petsko, G.A. and Ritz, J. (2020). Personalized
iPSC-Derived Dopamine Progenitor Cells for Parkinson’s Disease. New England
Journal of Medicine, 382(20), pp.1926–1932.
doi:https://doi.org/10.1056/nejmoa1915872.
63. Sebastiano, V., Zhen, H.H., Haddad, B., Derafshi, B.H., Bashkirova, E., Melo, S.P.,
Wang, P., Leung, T.L., Siprashvili, Z., Tichy, A., Li, J., Ameen, M., Hawkins, J., Lee,
S., Li, L., Schwertschkow, A., Bauer, G., Lisowski, L., Kay, M.A. and Kim, S.K.
(2014). Human COL7A1-corrected induced pluripotent stem cells for the treatment of
recessive dystrophic epidermolysis bullosa. Science Translational Medicine, [online]
6(264), p.264ra163. doi:https://doi.org/10.1126/scitranslmed.3009540.
64. Song, K., Nam, Y.-J., Luo, X., Qi, X., Tan, W., Huang, G.N., Acharya, A., Smith,
C.L., Tallquist, M.D., Neilson, E.G., Hill, J.A., Bassel-Duby, R. and Olson, E.N.
(2012). Heart repair by reprogramming non-myocytes with cardiac transcription
factors. Nature, 485(7400), pp.599–604. doi:https://doi.org/10.1038/nature11139.
65. Soufi, A., Donahue, G. and Zaret, Kenneth S. (2012). Facilitators and Impediments of
the Pluripotency Reprogramming Factors’ Initial Engagement with the Genome. Cell,
151(5), pp.994–1004. doi:https://doi.org/10.1016/j.cell.2012.09.045.
66. Stadtfeld, M. and Hochedlinger, K. (2010). Induced pluripotency: history,
mechanisms, and applications. Genes & Development, [online] 24(20), pp.2239–2263.
doi:https://doi.org/10.1101/gad.1963910.
67. Sugimoto, N., Kanda, J., Nakamura, S., Kitano, T., Hishizawa, M., Kondo, T.,
Shimizu, S., Shigemasa, A., Hirai, H., Arai, Y., Minami, M., Tada, H., Momose, D.,
Koh, K.-R., Nogawa, M., Watanabe, N., Okamoto, S., Handa, M., Sawaguchi, A. and
Matsuyama, N. (2022). iPLAT1: the first-in-human clinical trial of iPSC-derived
platelets as a phase 1 autologous transfusion study. Blood, 140(22), pp.2398–2402.
doi:https://doi.org/10.1182/blood.2022017296.
68. Sun, C., Serra, C., Lee, G. and Wagner, K.R. (2020). Stem cell-based therapies for
Duchenne muscular dystrophy. Experimental Neurology, 323, p.113086.
doi:https://doi.org/10.1016/j.expneurol.2019.113086.
69. Sun, H., Pratt, R.E., Dzau, V.J. and Hodgkinson, C.P. (2023). Neonatal and adult
cardiac fibroblasts exhibit inherent differences in cardiac regenerative capacity.
Journal of Biological Chemistry, 299(5), pp.104694–104694.
doi:https://doi.org/10.1016/j.jbc.2023.104694.
70. Takahashi, K. and Yamanaka, S. (2006). Induction of Pluripotent Stem Cells from
Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell, 126(4),
pp.663–676. doi:https://doi.org/10.1016/j.cell.2006.07.024.
71. Tanabe, K., Nakamura, M., Narita, M., Takahashi, K. and Yamanaka, S. (2013).
Maturation, not initiation, is the major roadblock during reprogramming toward
pluripotency from human fibroblasts. Proceedings of the National Academy of
Sciences, 110(30), pp.12172–12179. doi:https://doi.org/10.1073/pnas.1310291110.
72. Tang, S., Xie, M., Cao, N. and Ding, S. (2015). Patient-Specific Induced Pluripotent
Stem Cells for Disease Modeling and Phenotypic Drug Discovery. Journal of
Medicinal Chemistry, 59(1), pp.2–15.
doi:https://doi.org/10.1021/acs.jmedchem.5b00789.
73. Taylor, C.J., Bolton, E.M. and Bradley, J.A. (2011). Immunological considerations for
embryonic and induced pluripotent stem cell banking. Philosophical Transactions of
the Royal Society B: Biological Sciences, 366(1575), pp.2312–2322.
doi:https://doi.org/10.1098/rstb.2011.0030.
74. Wernig, M., Lengner, C.J., Hanna, J., Lodato, M.A., Steine, E., Foreman, R., Staerk,
J., Markoulaki, S. and Jaenisch, R. (2008). A drug-inducible transgenic system for
direct reprogramming of multiple somatic cell types. Nature Biotechnology, 26(8),
pp.916–924. doi:https://doi.org/10.1038/nbt1483.
75. White, M.C., Holman, D.M., Boehm, J.E., Peipins, L.A., Grossman, M. and Jane
Henley, S. (2014). Age and Cancer Risk. American Journal of Preventive Medicine,
[online] 46(3), pp.S7–S15. doi:https://doi.org/10.1016/j.amepre.2013.10.029.
76. Wiley, L.A., Burnight, E.R., DeLuca, A.P., Anfinson, K.R., Cranston, C.M., Kaalberg,
E.E., Penticoff, J.A., Affatigato, L.M., Mullins, R.F., Stone, E.M. and Tucker, B.A.
(2016). cGMP production of patient-specific iPSCs and photoreceptor precursor cells
to treat retinal degenerative blindness. Scientific Reports, 6(1).
doi:https://doi.org/10.1038/srep30742.
77. World Population Prospects, U. (2022). Life expectancy, 1770 to 2021. Our World in
Data. Available at: https://ourworldindata.org/grapher/life-expectancy?facet=none
78. Xia, M., Liu, K., Feng, J., Zheng, Z. and Xie, X. (2021). Prevalence and Risk Factors
of Type 2 Diabetes and Prediabetes Among 53,288 Middle-Aged and Elderly Adults
in China: A Cross-Sectional Study. Diabetes, Metabolic Syndrome and Obesity:
Targets and Therapy, Volume 14, pp.1975–1985.
doi:https://doi.org/10.2147/dmso.s305919.
79. Yang, J.-H., Petty, C.A., Dixon-McDougall, T., Maria Vina Lopez, Tyshkovskiy, A.,
Maybury-Lewis, S., Tian, X., Ibrahim, N., Chen, Z., Griffin, P.T., Arnold, M., Li, J.,
Martinez, O.A., Behn, A., Rogers-Hammond, R., Angeli, S., Gladyshev, V.N. and
Sinclair, D.A. (2023). Chemically induced reprogramming to reverse cellular aging.
Aging, 15(13). doi:https://doi.org/10.18632/aging.204896.
80. Yu, X., Abbas-Aghababazadeh, F., Chen, Y.A. and Fridley, B.L. (2020). Statistical
and Bioinformatics Analysis of Data from Bulk and Single-Cell RNA Sequencing
Experiments. Methods in Molecular Biology, 2194, pp.143–175.
doi:https://doi.org/10.1007/978-1-0716-0849-4_9.

Comments